Getting Started with pyGAM¶
Introduction¶
Generalized Additive Models (GAMs) are smooth semi-parametric models of the form:
where X.T = [X_1, X_2, ..., X_p] are independent variables, y is
the dependent variable, and g() is the link function that relates
our predictor variables to the expected value of the dependent variable.
The feature functions f_i() are built using penalized B splines,
which allow us to automatically model non-linear relationships
without having to manually try out many different transformations on
each variable.
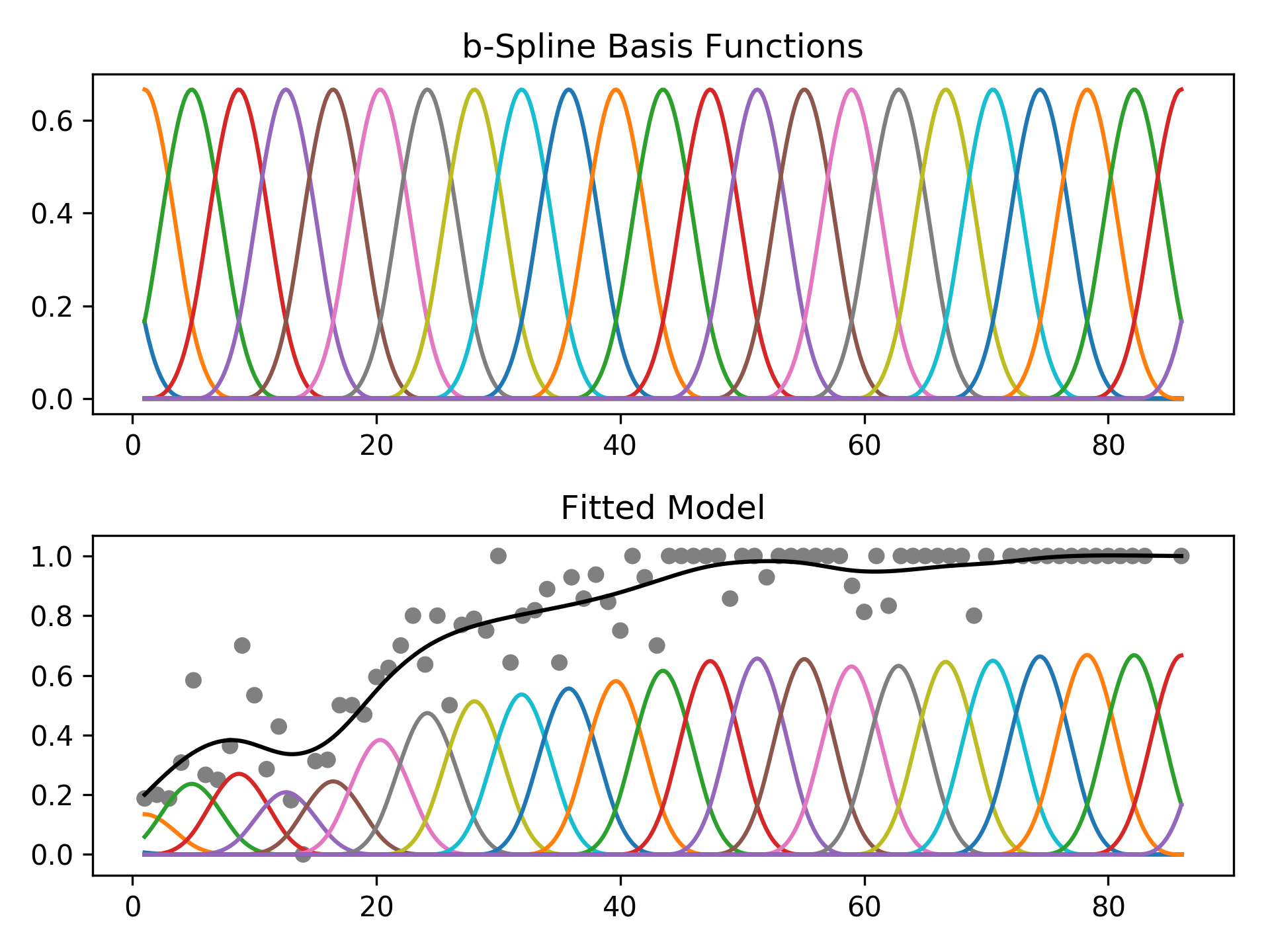
Basis splines
GAMs extend generalized linear models by allowing non-linear functions
of features while maintaining additivity. Since the model is additive,
it is easy to examine the effect of each X_i on Y individually
while holding all other predictors constant.
The result is a very flexible model, where it is easy to incorporate prior knowledge and control overfitting.
Regression¶
For regression problems, we can use a linear GAM which models:
In [1]:
import matplotlib.pyplot as plt
In [2]:
plt.ion()
plt.rcParams['figure.figsize'] = (12, 8)
In [3]:
from pygam import LinearGAM
from pygam.datasets import wage
X, y = wage(return_X_y=True)
gam = LinearGAM(n_splines=10).gridsearch(X, y)
XX = gam.generate_X_grid()
fig, axs = plt.subplots(1, 3)
titles = ['year', 'age', 'education']
for i, ax in enumerate(axs):
pdep, confi = gam.partial_dependence(XX, feature=i, width=.95)
ax.plot(XX[:, i], pdep)
ax.plot(XX[:, i], *confi, c='r', ls='--')
ax.set_title(titles[i])
100% (11 of 11) |########################| Elapsed Time: 0:00:01 Time: 0:00:01
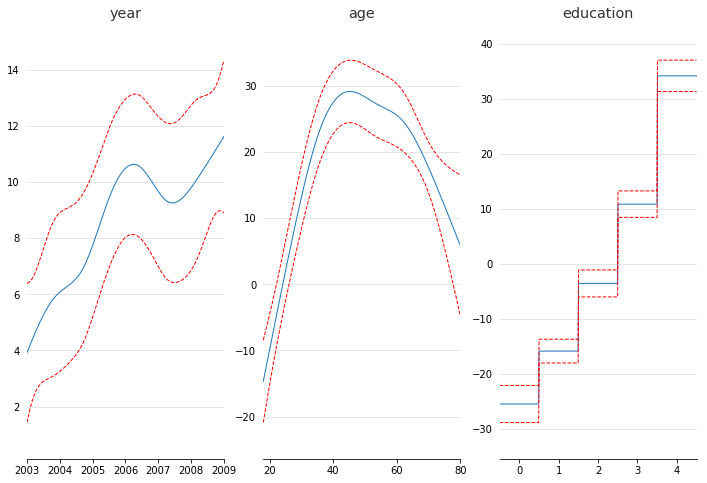
Even though we allowed n_splines=10 per numerical feature, our smoothing penalty reduces us to just 14 effective degrees of freedom:
In [4]:
gam.summary()
LinearGAM
=============================================== ==========================================================
Distribution: NormalDist Effective DoF: 13.532
Link Function: IdentityLink Log Likelihood: -24119.2327
Number of Samples: 3000 AIC: 48267.5293
AICc: 48267.6805
GCV: 1247.0706
Scale: 1236.9495
Pseudo R-Squared: 0.2926
==========================================================================================================
Feature Function Data Type Num Splines Spline Order Linear Fit Lambda P > x Sig. Code
================== ============== ============= ============= =========== ========== ========== ==========
feature 1 numerical 10 3 False 15.8489 8.23e-03 **
feature 2 numerical 10 3 False 15.8489 1.11e-16 ***
feature 3 categorical 5 0 False 15.8489 1.11e-16 ***
intercept 1.11e-16 ***
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
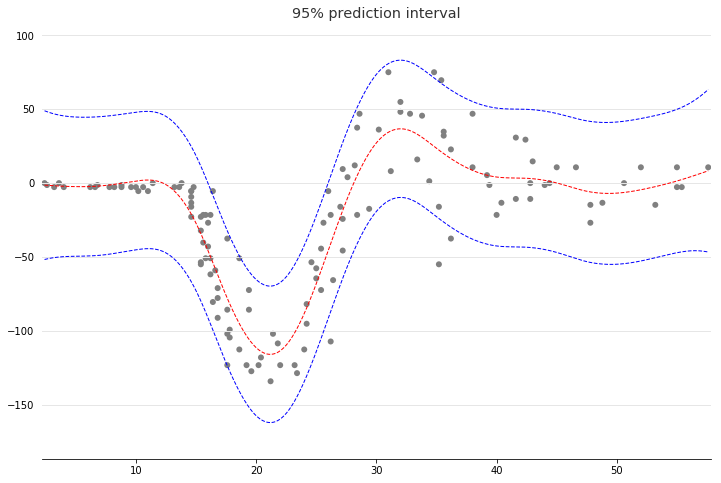
With LinearGAMs, we can also check the prediction intervals:
In [5]:
from pygam.datasets import mcycle
X, y = mcycle(return_X_y=True)
gam = LinearGAM().gridsearch(X, y)
XX = gam.generate_X_grid()
plt.plot(XX, gam.predict(XX), 'r--')
plt.plot(XX, gam.prediction_intervals(XX, width=.95), color='b', ls='--')
plt.scatter(X, y, facecolor='gray', edgecolors='none')
plt.title('95% prediction interval')
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[5]:
Text(0.5,1,'95% prediction interval')
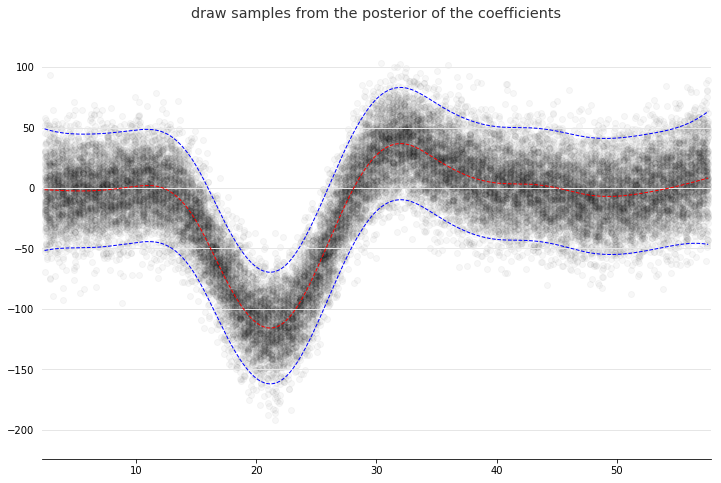
And simulate from the posterior:
In [6]:
# continuing last example with the mcycle dataset
for response in gam.sample(X, y, quantity='y', n_draws=50, sample_at_X=XX):
plt.scatter(XX, response, alpha=.03, color='k')
plt.plot(XX, gam.predict(XX), 'r--')
plt.plot(XX, gam.prediction_intervals(XX, width=.95), color='b', ls='--')
plt.title('draw samples from the posterior of the coefficients')
Out[6]:
Text(0.5,1,'draw samples from the posterior of the coefficients')
Classification¶
For binary classification problems, we can use a logistic GAM which models:
In [7]:
from pygam import LogisticGAM
from pygam.datasets import default
X, y = default(return_X_y=True)
gam = LogisticGAM().gridsearch(X, y)
XX = gam.generate_X_grid()
fig, axs = plt.subplots(1, 3)
titles = ['student', 'balance', 'income']
for i, ax in enumerate(axs):
pdep, confi = gam.partial_dependence(XX, feature=i, width=.95)
ax.plot(XX[:, i], pdep)
ax.plot(XX[:, i], confi[0], c='r', ls='--')
ax.set_title(titles[i])
100% (11 of 11) |########################| Elapsed Time: 0:00:07 Time: 0:00:07
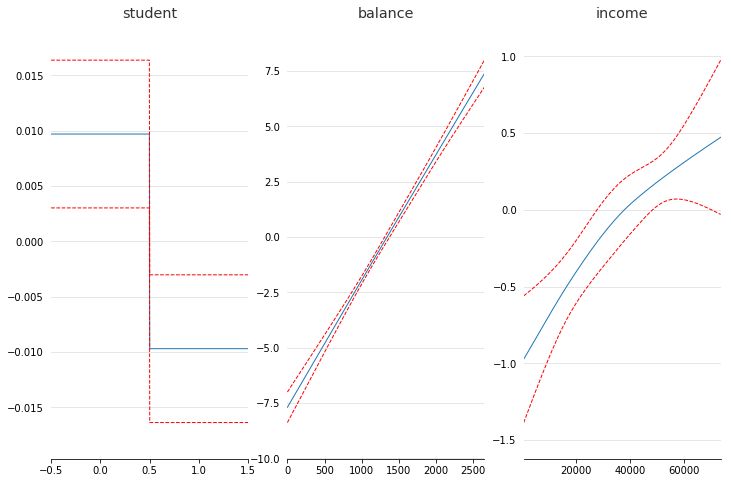
We can then check the accuracy:
In [8]:
gam.accuracy(X, y)
Out[8]:
0.9739
Since the scale of the Binomial distribution is known, our gridsearch minimizes the Un-Biased Risk Estimator (UBRE) objective:
In [9]:
gam.summary()
LogisticGAM
=============================================== ==========================================================
Distribution: BinomialDist Effective DoF: 4.3643
Link Function: LogitLink Log Likelihood: -788.7121
Number of Samples: 10000 AIC: 1586.1527
AICc: 1586.1595
UBRE: 2.159
Scale: 1.0
Pseudo R-Squared: 0.4599
==========================================================================================================
Feature Function Data Type Num Splines Spline Order Linear Fit Lambda P > x Sig. Code
================== ============== ============= ============= =========== ========== ========== ==========
feature 1 categorical 2 0 False 1000.0 4.41e-03 **
feature 2 numerical 25 3 False 1000.0 0.00e+00 ***
feature 3 numerical 25 3 False 1000.0 2.35e-02 *
intercept 0.00e+00 ***
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
Poisson and Histogram Smoothing¶
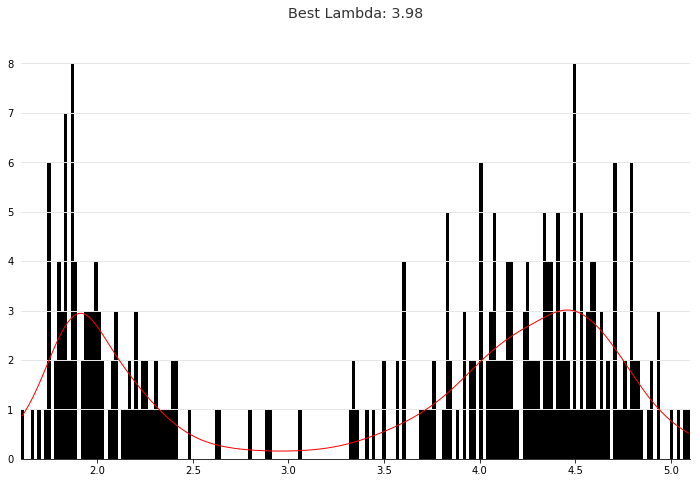
We can intuitively perform histogram smoothing by modeling the counts in each bin as being distributed Poisson via PoissonGAM.
In [10]:
from pygam import PoissonGAM
from pygam.datasets import faithful
X, y = faithful(return_X_y=True)
gam = PoissonGAM().gridsearch(X, y)
plt.hist(faithful(return_X_y=False)['eruptions'], bins=200, color='k');
plt.plot(X, gam.predict(X), color='r')
plt.title('Best Lambda: {0:.2f}'.format(gam.lam))
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[10]:
Text(0.5,1,'Best Lambda: 3.98')
Custom Models¶
It’s also easy to build custom models, by using the base GAM class and specifying the distribution and the link function.`
In [11]:
from pygam import GAM
from pygam.datasets import trees
X, y = trees(return_X_y=True)
gam = GAM(distribution='gamma', link='log', n_splines=4)
gam.gridsearch(X, y)
plt.scatter(y, gam.predict(X))
plt.xlabel('true volume')
plt.ylabel('predicted volume')
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[11]:
Text(0,0.5,'predicted volume')
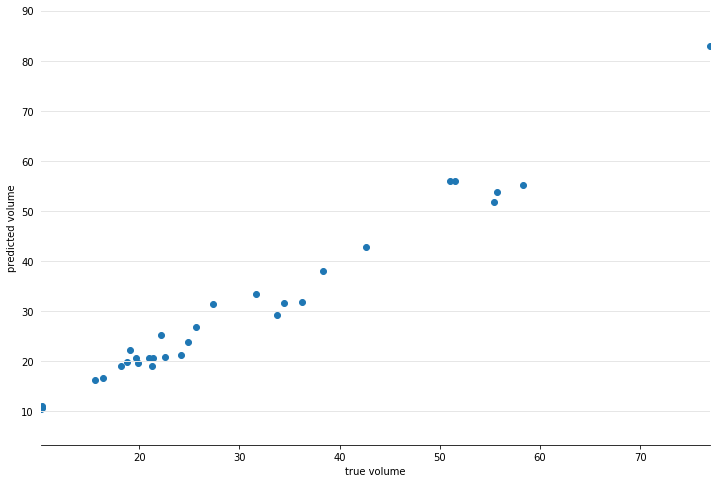
We can check the quality of the fit by looking at the Pseudo R-Squared:
In [12]:
gam.summary()
GAM
=============================================== ==========================================================
Distribution: GammaDist Effective DoF: 4.1544
Link Function: LogLink Log Likelihood: -66.9372
Number of Samples: 31 AIC: 144.1832
AICc: 146.7368
GCV: 0.0095
Scale: 0.0073
Pseudo R-Squared: 0.9767
==========================================================================================================
Feature Function Data Type Num Splines Spline Order Linear Fit Lambda P > x Sig. Code
================== ============== ============= ============= =========== ========== ========== ==========
feature 1 numerical 4 3 False 0.0158 1.11e-16 ***
feature 2 numerical 4 3 False 0.0158 8.28e-05 ***
intercept 1.11e-16 ***
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
Penalties / Constraints¶
With GAMs we can encode prior knowledge and control overfitting by using penalties and constraints.
Available penalties - second derivative smoothing (default on numerical features) - L2 smoothing (default on categorical features)
Availabe constraints - monotonic increasing/decreasing smoothing - convex/concave smoothing - periodic smoothing [soon…]
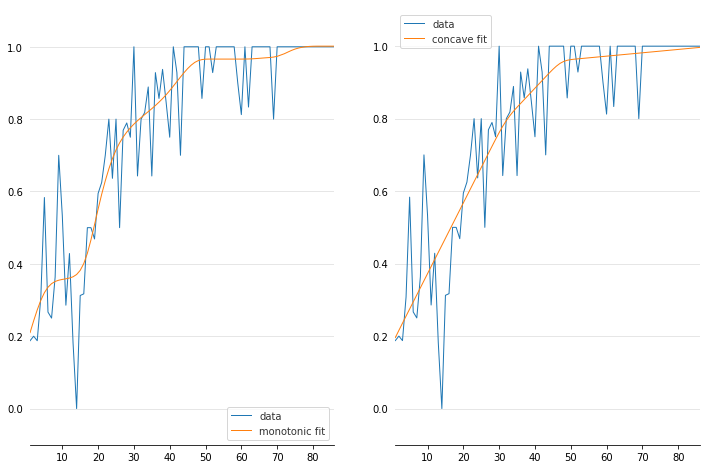
We can inject our intuition into our model by using monotonic and concave constraints:
In [13]:
from pygam import LinearGAM
from pygam.datasets import hepatitis
X, y = hepatitis(return_X_y=True)
gam1 = LinearGAM(constraints='monotonic_inc').fit(X, y)
gam2 = LinearGAM(constraints='concave').fit(X, y)
fig, ax = plt.subplots(1, 2)
ax[0].plot(X, y, label='data')
ax[0].plot(X, gam1.predict(X), label='monotonic fit')
ax[0].legend()
ax[1].plot(X, y, label='data')
ax[1].plot(X, gam2.predict(X), label='concave fit')
ax[1].legend()
Out[13]:
<matplotlib.legend.Legend at 0xd9c8550>
API¶
pyGAM is intuitive, modular, and adheres to a familiar API:
In [14]:
from pygam import LogisticGAM
from pygam.datasets import toy_classification
X, y = toy_classification(return_X_y=True, n=5000)
gam = LogisticGAM()
gam.fit(X, y)
Out[14]:
LogisticGAM(callbacks=[Deviance(), Diffs(), Accuracy()],
constraints=None, dtype='auto', fit_intercept=True,
fit_linear=False, fit_splines=True, lam=0.6, max_iter=100,
n_splines=25, penalties='auto', spline_order=3, tol=0.0001,
verbose=False)
Since GAMs are additive, it is also super easy to visualize each
individual feature function, f_i(X_i). These feature functions
describe the effect of each X_i on y individually while
marginalizing out all other predictors:
In [15]:
import numpy as np
pdeps = gam.partial_dependence(np.sort(X, axis=0))
_ = plt.plot(pdeps)
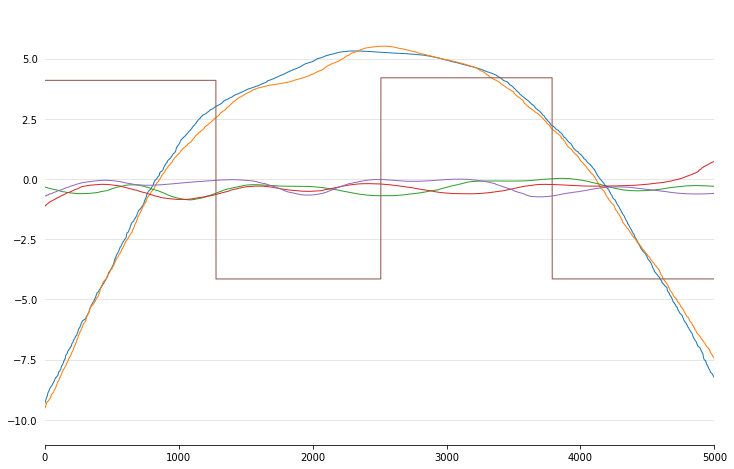
Current Features¶
Models¶
pyGAM comes with many models out-of-the-box:
- GAM (base class for constructing custom models)
- LinearGAM
- LogisticGAM
- GammaGAM
- PoissonGAM
- InvGaussGAM
You can mix and match distributions with link functions to create custom models!
In [16]:
GAM(distribution='gamma', link='inverse')
Out[16]:
GAM(callbacks=['deviance', 'diffs'], constraints=None,
distribution='gamma', dtype='auto', fit_intercept=True,
fit_linear=False, fit_splines=True, lam=0.6, link='inverse',
max_iter=100, n_splines=25, penalties='auto', spline_order=3,
tol=0.0001, verbose=False)
Distributions¶
- Normal
- Binomial
- Gamma
- Poisson
- Inverse Gaussian
Link Functions¶
Link functions take the distribution mean to the linear prediction. These are the canonical link functions for the above distributions:
- Identity
- Logit
- Inverse
- Log
- Inverse-squared
Callbacks¶
Callbacks are performed during each optimization iteration. It’s also easy to write your own.
- deviance - model deviance
- diffs - differences of coefficient norm
- accuracy - model accuracy for LogisticGAM
- coef - coefficient logging
You can check a callback by inspecting:
In [17]:
_ = plt.plot(gam.logs_['deviance'])
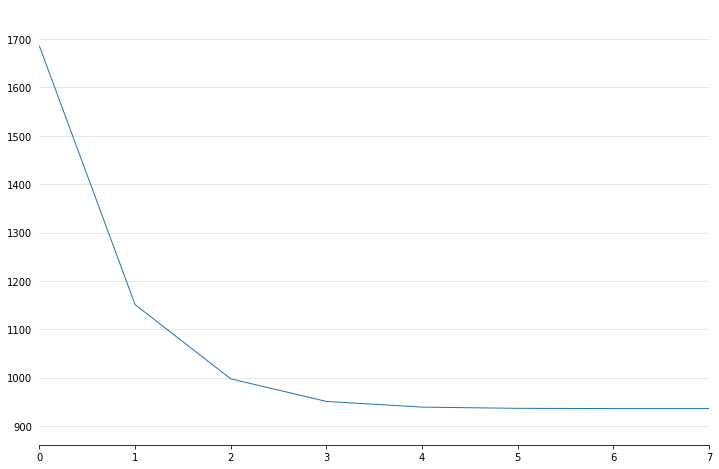
Linear Extrapolation¶
In [18]:
X, y = mcycle()
gam = LinearGAM()
gam.gridsearch(X, y)
XX = gam.generate_X_grid()
m = X.min()
M = X.max()
XX = np.linspace(m - 10, M + 10, 500)
Xl = np.linspace(m - 10, m, 50)
Xr = np.linspace(M, M + 10, 50)
plt.figure()
plt.plot(XX, gam.predict(XX), 'k')
plt.plot(Xl, gam.confidence_intervals(Xl), color='b', ls='--')
plt.plot(Xr, gam.confidence_intervals(Xr), color='b', ls='--')
_ = plt.plot(X, gam.confidence_intervals(X), color='r', ls='--')
100% (11 of 11) |########################| Elapsed Time: 0:00:00 Time: 0:00:00
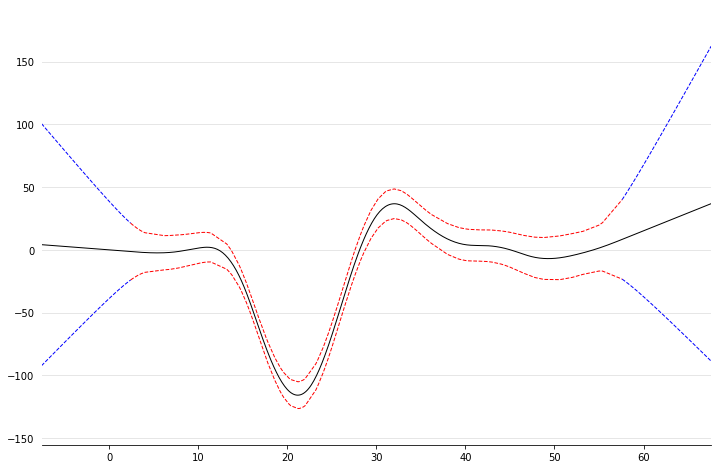
Citing pyGAM¶
Please consider citing pyGAM if it has helped you in your research or work:
Daniel Servén, & Charlie Brummitt. (2018, March 27). pyGAM: Generalized Additive Models in Python. Zenodo. DOI: 10.5281/zenodo.1208723
BibTex:
@misc{daniel\_serven\_2018_1208723,
author = {Daniel Servén and
Charlie Brummitt},
title = {pyGAM: Generalized Additive Models in Python},
month = mar,
year = 2018,
doi = {10.5281/zenodo.1208723},
url = {https://doi.org/10.5281/zenodo.1208723}
}
References¶
- Simon N. Wood, 2006Generalized Additive Models: an introduction with R
- Hastie, Tibshirani, FriedmanThe Elements of Statistical Learning
- James, Witten, Hastie and TibshiraniAn Introduction to Statistical Learning
Paul Eilers & Brian Marx, 1996 Flexible Smoothing with B-splines and Penalties http://www.stat.washington.edu/courses/stat527/s13/readings/EilersMarx_StatSci_1996.pdf
- Kim Larsen, 2015GAM: The Predictive Modeling Silver Bullet
- Deva Ramanan, 2008UCI Machine Learning: Notes on IRLS
- Paul Eilers & Brian Marx, 2015International Biometric Society: A Crash Course on P-splines
- Keiding, Niels, 1991Age-specific incidence and prevalence: a statistical perspective